import numpy as np
import pandas as pd
import xml.etree.ElementTree as ET
from os import walk
_, _, filenames = next(walk('news'))
df = pd.DataFrame()
for file in filenames:
xml = ET.parse("news/" + file).getroot()
for entry in xml.findall('entry'):
title= entry.find("title").text
text = entry.find("text").text
site = entry.find("link").text
tag = entry.find("tag").text
df = df.append({"title": title, "text": text, "tag": tag, "site": site}, ignore_index=True)
df.head()NLP: Clustering Inception
When using supervised learning for classifying data you normally begin with some labelled dataset. These labels help in the evaluation phase model to assess how probable it is that a given sample belongs to a specific target class.
But sometimes you’d like to classify unlabelled data. That usually suggests the use of unsupervised learning techniques such as clustering or grouping data by some common feature. Then imagine you’ve got a bunch of unclassified text documents and get some groups by clustering and finally use those groups to classify new documents. That would be great.
In this article I’m going all the way and take a list of unlabelled texts, clustering them into groups and finally try to classify new documents based on these groups or clusters.
Loading Data
I’m loading some news articles from different sources and about different topics to create a Pandas DataFrame. Although the data has labels, I’m only using them at the end to check how well the whole process worked.

Cleaning Data
In this section, I’m going to normalize the text. There are several tasks that people normally do to normalize data, such getting rid of stop-words, removing extra white spaces…etc. A couple of weeks ago I heard about TextHero which among other things it provides a solid pipeline to clean data. With TextHero I can very easily do some basic cleaning:
-
Replace not assigned values with empty spaces.
-
Lowercase all text.
-
Remove all blocks of digits.
-
Remove all string.punctuation (!"#$%&'()*+,-./:;<⇒?@[\]^_`{|}~).
-
Remove all accents from strings.
-
Remove all stop words.
-
Remove all white space between words.
Apart from the basic cleaning tasks I needed to remove some stuff that I think could be considered as data leakage such as:
-
The web site of the article
-
The topic the newspaper grouped the article
-
some other words that I could consider noise
# creating a copy of source dataset
articles_df = df.copy()
# article topics
topics = articles_df['tag'].unique()
# some words under my consideration
words_to_delete = ['said', 'also', 'year', 'would']
# sites of the articles
sites = articles_df['site']\
.str.extractall(r'https://.*\.(?P<www>\w*)\..*')\
.reset_index(col_fill='origin')['www']\
.unique()The nice thing about TextHero is that I can create a pipeline of different functions that will be applied to every sample to clean the data:
import texthero as hero
from texthero import preprocessing as pre
# custom pipeline
custom_pipeline = [
# default cleaning functions
pre.fillna,
pre.lowercase,
pre.remove_digits,
pre.remove_punctuation,
pre.remove_diacritics,
pre.remove_stopwords,
pre.remove_whitespace,
# extra cleaning tasks
remove(topics),
remove(sites),
remove(words_to_delete)
]
# cleaning text with custom pipeline
articles_df['text'] = articles_df['text'].pipe(hero.clean, custom_pipeline)
articles_df['text'].head()
Here’s the source of the remove(…) function:
from functools import reduce
def remove(words):
def remove_from_series(s):
return reduce(lambda acc, val: acc.str.replace(val, ""), words, s)
return remove_from_seriesSplitting Data
Ok, I’ve got my data cleaned, or at least, sort of. It’s time to split the data in two datasets:
-
Half of the data will be used for training the clustering model
-
The other half will be used for testing the classifier created from the clustering model
articles_df['business'] = np.where(articles_df['tag'] == 'business', 1, 0)
groups = articles_df.groupby('business')
busi = groups.get_group(1)
busi_half = round(len(busi) / 2)
non_busi = groups.get_group(0)
non_busi_half = round(len(non_busi) / 2)
train = pd\
.concat([busi[:busi_half], non_busi[:non_busi_half]])\
.sample(frac=1)\
.reset_index(drop=True)
tests = pd\
.concat([busi[busi_half:], non_busi[non_busi_half:]])\
.sample(frac=1)\
.reset_index(drop=True)
len(train), len(tests)(478, 479)Clustering
Before asking the model to group all news into two topics, I need to:
-
Transform text to something that the model can work with
-
Create a corpus
-
Create a dictionary
import gensim
from sklearn.feature_extraction.text import CountVectorizer
txt = train['text']
vec = CountVectorizer()
X = vec.fit_transform(txt)
# Convert sparse matrix to gensim corpus.
corpus = gensim.matutils.Sparse2Corpus(X, documents_columns=False)
# Mapping from word IDs to words (To be used in LdaModel's id2word parameter)
dictionary = dict((v, k) for k, v in vec.vocabulary_.items())LdaModel
To create the clustering model I’m using Gensim and its LDA model implementation. According to Gensim’s documentation: This module allows both LDA model estimation from a training corpus and inference of topic distribution on new, unseen documents. The model can also be updated with new documents for online training.
# Use the gensim.models.ldamodel.LdaModel constructor to estimate
# LDA model parameters on the corpus, and save to the variable `ldamodel`
from gensim.models.ldamodel import LdaModel
# Your code here:
ldamodel = LdaModel(corpus, id2word=id_map, num_topics=2)Invoking the model’s show_topics function will show us the two topics found and the terms or tokens related to them with their weight. In other words, how important these terms are to consider that a given article belongs to that topic.
ldamodel.show_topics()[(0,
'0.004*"biden" + 0.004*"last" + 0.004*"new" + 0.003*"one" + 0.003*"like" + 0.003*"people" + 0.003*"president" + ...'),
(1,
'0.004*"first" + 0.003*"mpany" + 0.003*"new" + 0.003*"one" + 0.003*"back" + 0.003*"people" + 0.003*"get" + 0.003...')]So if a given article has biden or president is more likely to belong to the topic 0 whereas if another article has company is more likely to belong to the topic 1. At this point we could manually label those topics, for example, topic 0 could become non_business and topic 1 could become business, and then just use those labels in a binary classification.
Binary classification using LDA model
The cluster model created two topics, which means that we can label samples by looking at the distribution of the topics for each document. Because we only have two topics If the distribution of a given topic is greater than the other then I’ll assume it belongs to that topic. We can get the distribution of topics for a given document by invoking get_document_topics(…) from the LDA model we’ve created previously.
This way I’ve created a labelled dataset from a bunch of unlabelled articles and now I’d like to use this new labelled dataset to classify the other half of the articles we left for testing purposes.
# transforming raw data
X = vec.transform(tests['text'])
# creating a new corpus to cluster
corpus = gensim.matutils.Sparse2Corpus(X, documents_columns=False)
# creating a series to compare results with initial labels
results = []
for next_doc in ldamodel.get_document_topics(corpus):
if len(next_doc) == 2:
_, pct_zero = next_doc[0]
_, pct_one = next_doc[1]
results.append(1 if pct_one > pct_zero else 0)
else:
target, pct = next_doc[0]
results.append(target)
check = tests.copy()
check['classified'] = pd.Series(results)
check.loc[10:20, ['title', 'business', 'classified']]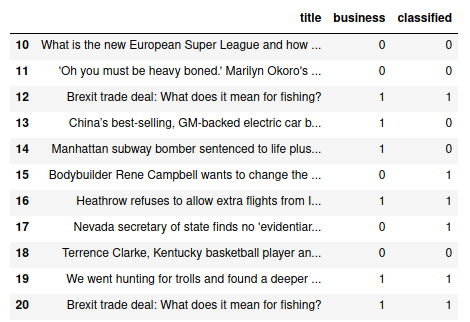
Well just by looking at the picture, it doesn’t seem great, but, How accurate is how classification model in reality ? Lets figure out the accuracy score with sklearn’s accuracy_score(…) function:
from sklearn.metrics import accuracy_score
accuracy_score(check['business'], check['classified'])0.732776617954071Well, more than I expected.